What Do We Know About the New Variant of Coronavirus?
The talk on everyone’s lips this week is the new Covid variant identified in the the UK. Concern surrounding the new strain has sapped some of the elation brought on by the first rollout of vaccines. While there’s no evidence so far that the UK variant — or any of the other known variants — has any impact on the efficacy of vaccines, early reports suggest the new kid on the block is well worth paying attention to, as its more contagious nature threatens to test our resolve over the next few months and push our already overburdened health care systems to their limit.
Here’s the original paper on the SARS-2 variant known as B.1.1.7 that emerged in the UK at least as early as September. One question that’s on a lot of people’s minds is: how did this happen? The received wisdom was that SARS-2, like its coronavirus kin, is relatively genetically stable in both humans and animals, certainly when compared to influenza and other RNA-based viruses. (It’s why we develop a new flu vax every year, but we don’t for measles.) For example, Carl Zimmer wrote the following on the 30th of April (emphasis mine):
“In fact, researchers have found that the coronavirus is mutating relatively slowly compared to some other RNA viruses, in part because virus proteins acting as proofreaders are able to fix some mistakes. Each month, a lineage of coronaviruses might acquire only two single-letter mutations.
In the future, the coronavirus may pick up some mutations that help it evade our immune systems. But the slow mutation rate of the coronavirus means that these changes will emerge over the course of years.”
Of course, this never meant that the novel SARS-CoV-2 wouldn’t defy our expectations (as it has in several other respects), or that science doesn’t change. More people getting sick means more replicating viruses. More replication means more mutations. And more mutations mean more opportunities for the coronavirus to change in significant, even dangerous ways. The longevity of the pandemic, and the fact that SARS-2 has managed to worm its way into nearly every corner of the globe, raised the prospect of new strains emerging, and here they are — if a bit sooner than expected.
What appears to have happened, according to the running hypothesis in the above paper, is that the UK strain originated from a prolonged infection in an immunocompromised host. Coronaviruses isolated from patients with prolonged or persistent bouts of Covid show an unusually high amount of genetic variation. These patients tend to be immunodeficient in some way or another, which biases them toward chronic infection as the virus is allowed to linger in the body for a much longer period of time compared to people with healthy immune systems.
And it’s precisely this combination that produced the unique evolutionary pressures that gave rise to the UK strain:
Weak selective pressure from the patient’s natural immune response
⬇
Unusually large and genetically diverse virus population that builds up over the course of the infection
⬇
Strong selective pressure after convalescent plasma or other forms of antibody therapy are introduced several weeks later
In short, hosts with a weakened immune system foster a higher degree of viral evolution compared to immunocompetent individuals due to some unique factors at play in such hosts.
The authors note that patients with this particular type of infection are a rarity, and that onward transmission from said patients should be rarer still. Patients who develop chronic infections are usually hospitalized and under the care and supervision of trained health care workers equipped with proper PPE. Thus forward transmission is unlikely, or at least far less likely compared to asymptomatic “silent spreaders” walking around with no clue they’re even carrying the virus. In this case, however, transmission did occur. It sounds like we may have just gotten unlucky. It may be that the new strain’s more contagious properties managed to crash through the normal impediments.
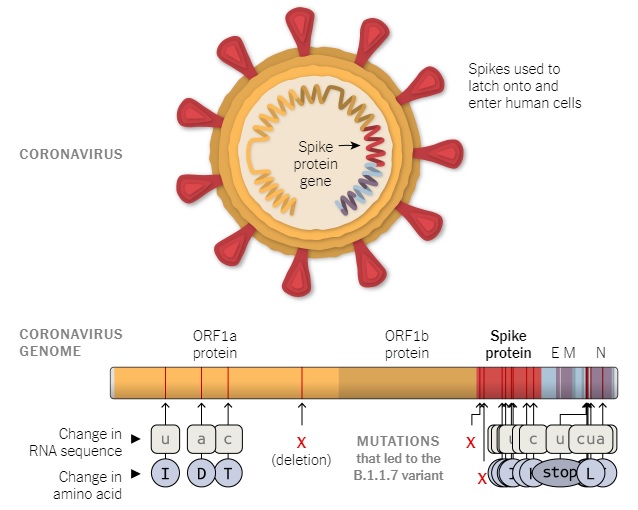
The researchers identified a total of 23 mutations present in the new variant, including 6 synonymous mutations, 14 non-synonymous mutations, and 3 deletions.
According to a preprint released by British scientists this past Wednesday, the cumulative effect of these various mutations is more rapid spread. While the variant is not currently linked to more severe illness or higher risk of hospitalization or death, initial estimates indicate B.1.1.7 is between 56 and 70 percent more infectious compared to others that have been circulating in most parts of the world. That’s alarming news for nations already seeing record high case counts and deaths as we settle in for the winter and outdoor activity wanes.
From the moment the new variant was announced, many experts suggested it was essentially a foregone conclusion that it wouldn’t stay confined to the UK for long, and likely had already escaped. Given the shamefully low rates of testing across the US in particular, the lack of a national program for genetic sequencing, and the reality of international travel, one could easily presume the mutant strain had been circulating here for weeks, if not months, without detection.
Sure enough, news came this morning that the first case of B.1.1.7 in the US has been confirmed in Colorado. More than a dozen other countries have also reported finding the new variant in samples isolated from patients. That the variant had been on hand but not accounted for was bolstered by the fact that the patient in Colorado had no travel history. Presumably, it was spreading among the community for quite some time.
One silver lining in all this is that most scientists doubt the new strains will undermine the vaccine effort. The CDC is currently tracking a number of SARS-2 variants or strains1 identified over the last few months, one from the UK (B.1.1.7) and one in South Africa (B.1.351). Under both, the agency emphasizes that there is no evidence either variant “has any impact on disease severity or vaccine efficacy.” Dr. Fauci appears optimistic as well. That said, we cannot yet rule out the possibility, and a number of countries and vaccine makers are hustling to secure more concrete answers to this very question.
The scientists behind the B.1.1.7 paper do close out on a hopeful note by reflecting on a strain that emerged in Singapore. That strain was discovered earlier this year and produced a milder form of Covid in patients. However, it died out shortly afterward thanks to the swift control measures enacted by the government. Singapore’s encounter with its own unique variant can serve as a hopeful lesson for Western nations by demonstrating that similar successes can be achieved if we act quickly and decisively.
The revelation of a new, faster-spreading variant comes at a most inopportune time. Just as the world begins to celebrate the influx of vaccines, a more formidable foe has arrived that threatens to complicate our ability to control the virus. But there is also an opportunity here. The emergence of new strains sharpens the urgency of vaccination programs around the world. We — from global health agencies to ordinary citizens — can respond to these new perils in the months ahead by stepping up surveillance and testing, and by redoubling our efforts to distribute the vaccines as quickly, as widely, and as safely as possible. First and foremost, these efforts must entail educating people about vaccine safety and the role we all play in minimizing transmission and bringing the virus to heel.
As it’s often said, vaccines don’t save lives, vaccination campaigns do.
Further reading:
- What we know about the new British variant.
- U.K. variant puts spotlight on immunocompromised patients’ role in the COVID-19 pandemic
- Coronavirus Variant Is Indeed More Transmissible, New Study Suggests
- First U.S. Case of Highly Contagious Coronavirus Variant Is Found in Colorado
- CEPI creates new collaborative taskforce to assess impact of emerging viral strains on effectiveness of COVID-19 vaccines
- Vaccine makers in Asia rush to test jabs against fast-spreading COVID variant
- The new U.K. coronavirus variant is concerning. But don’t freak out
- Could too much time between doses drive the coronavirus to outwit vaccines?
- Inside the B.1.1.7 Coronavirus Variant
- COVID Variants May Arise in People with Compromised Immune Systems (study)
Image credit: Shutterstock
- Virology can be a notoriously confusing field both because the terminology used in the literature is often inconsistent, and because popular concepts within the field are still fiercely debated. (Welcome to academia.) As an example, terms like ‘isolate,’ ‘strain,’ ‘variant,’ ‘species,’ and ‘serotype’ are not always used the same way by different infectious disease researchers; while some associate these terms with different concepts, others may use them more or less interchangeably, or worse, inconsistently.
The CDC weighed in on this matter in their interim report on B.1.1.7: “The press often uses the terms “variant,” “strain,” “lineage,” and “mutant” interchangeably. For the time being in the context of this variant, the first three of these terms are generally being used interchangeably by the scientific community as well.”
Typically, the terms mentioned by the CDC above are reserved for versions with special changes that confer a new property to the virus. Since lineage B.1.1.7 appears to be up to 70% more transmissible thanks to a few key mutations, it’s acceptable to refer to it as a new ‘strain’ or ‘variant.’ Other viral genomes isolated from patients, which number in the hundreds of thousands at this point, have no new or discerning features or properties to write home about, and are merely considered ‘isolates.’
To be sure, virologists will not necessarily agree on this, just as many did not agree with referring to the Singapore virus as a ‘new strain.’ The conversation will go on. [↩]


Comments